Peer-reviewed Paper
We describe the application of Intellegens’ unique deep learning technology to the drug discovery domain. In two test case studies on public domain data sets we show that the method is able to accurately predict protein activity levels, substantially outperforming leading computational chemistry methods.
We have presented a new neural network imputation technique for predicting bioactivity, which can learn from incomplete bioactivity data to improve the quality of predictions by using correlations between both different bioactivity assays.
Abstract
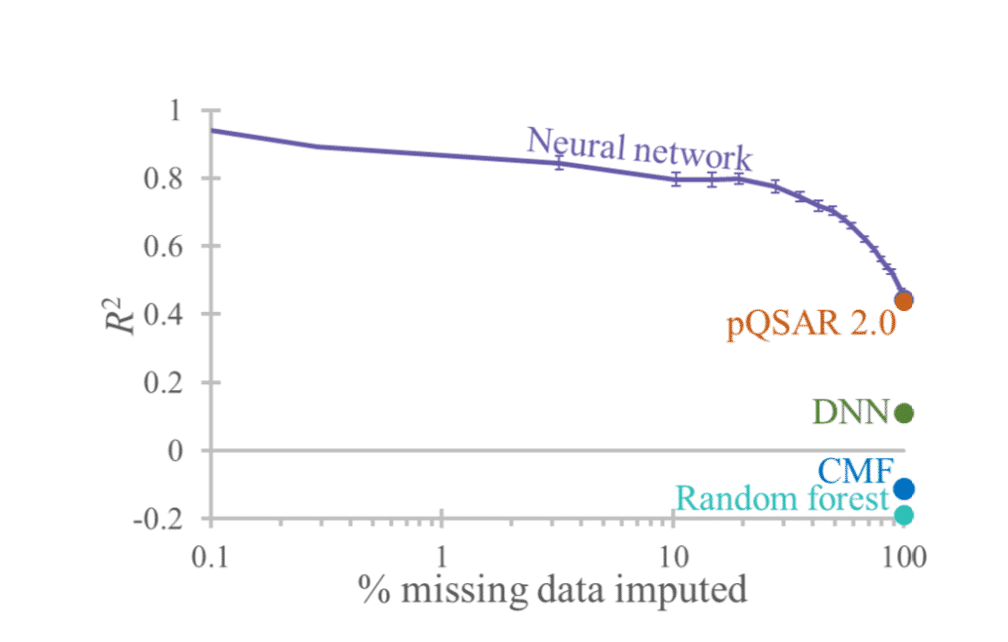
We describe a novel deep learning neural network method and its application to impute assay pIC50 values. Unlike conventional machine learning approaches, this method is trained on sparse bioactivity data as input, typical of that found in public and commercial databases, enabling it to learn directly from correlations between activities measured in different assays. In two case studies on public domain data sets we show that the neural network method outperforms traditional quantitative structure-activity relationship (QSAR) models and other leading approaches. Furthermore, by focussing on only the most confident predictions the accuracy is increased to R2 > 0.9 using our method, as compared to R2 = 0.44 when reporting all predictions.
Publication details
Journal: J. Chem. Inf. Model.
Title: Imputation of Assay Bioactivity Data Using Deep Learning
Authors: T. M. Whitehead*, B. W. J. Irwin, P. Hunt, M. D. Segall, and G. J. Conduit
DOI / Link: https://doi.org/10.1021/acs.jcim.8b00768