Oligonucleotides are a hot area of life science research, offering huge therapeutic potential. But such benefits rarely come easily. For oligonucleotides, this is perhaps most obvious in the difficulties of achieving reliable production processes. In our latest blog, we explore why oligonucleotides are important, some key development challenges, and how machine learning (ML) is now leveraging the increasing pool of available process data to help maximize yields and minimize impurities.

Oligonucleotides are single-stranded chains of nucleotides – the fundamental building blocks of DNA and RNA. These molecules can be custom-built with any sequence we choose, which means we can design oligonucleotides to precisely interact with and influence gene expression inside cells, allowing us to influence or correct gene expression for therapeutic purposes. This paves the way for breakthroughs in personalized medicine, potentially tackling conditions from rare genetic disorders to complex cancers.
However, translating that precision from design to production is far from straightforward. Despite their relatively short length, oligonucleotides are chemically complex molecules, and their synthesis involves dozens of interdependent steps, each one a potential source of yield loss or impurity formation. Inefficient coupling reactions, incomplete deprotection, side reactions from reactive intermediates, moisture sensitivity, reagent degradation, and purification challenges all contribute to a significant accumulation of impurities and variable yields. The balance between reaction efficiency, reagent excess, and process conditions such as temperature, solvent, and timing is delicate. Even minor deviations can lead to measurable losses in product quality and quantity. As a result, the synthesis of oligonucleotides remains highly dependent on expert intuition and iterative experimentation. Process optimization often relies on a small group of specialists capable of interpreting complex analytical data and adjusting parameters based on experience. This expert-driven approach, while effective in isolated cases, is difficult to scale and limits broader process reproducibility.
This is where machine learning (ML) offers transformative potential. The synthesis process generates large volumes of experimental and analytical data—reaction conditions, sequence characteristics, chromatographic profiles, impurity spectra, and more. Within this data lies the key to understanding how specific process parameters influence impurity formation and yield outcomes. ML can uncover these hidden relationships, identifying which combinations of parameters consistently lead to cleaner synthesis and higher efficiency.
Can’t we just build a machine learning model and use it to predict, for example, which parameters will maximize yields and minimize impurities for a given oligonucleotide sequence? The answer is yes, we can – although this process has its own challenges.
By training models to predict impurity patterns, optimal coupling conditions, or purification success based on both numerical and sequence-derived data, ML can complement the expertise of chemists by uncovering subtle data-driven relationships that inform parameter selection and streamline experimental optimization. Over time, such systems could move from retrospective analysis to real-time optimization, helping chemists anticipate impurity risks before they occur and enabling them to achieve higher yields with fewer iterations. But this process requires a framework and ML technology that can bring together diverse data sources, make it straightforward to organize the data for input to ML models, and apply robust models that generate useful results even from imperfect or partial data, giving clear guidance on the uncertainty in the predictions. Tackling these challenges has been the focus of a two-year project led by Intellegens in collaboration with CPI and six major pharmaceutical and biotechnology companies.
Together, the team has developed a software tool that makes it easy to bring together raw synthesis logs, deconvoluted mass spectrometry outputs, and reagent/yield data. Using the Alchemite™ method, the tool builds a machine learning model capable of providing sequence-specific recommendations for process parameters that will reduce the impurity burden, improve crude yield, and cut the number of experiments required. Such guidance reduces the need for senior expert input on more routine aspects of process development, so that valuable resources can be better-focused.
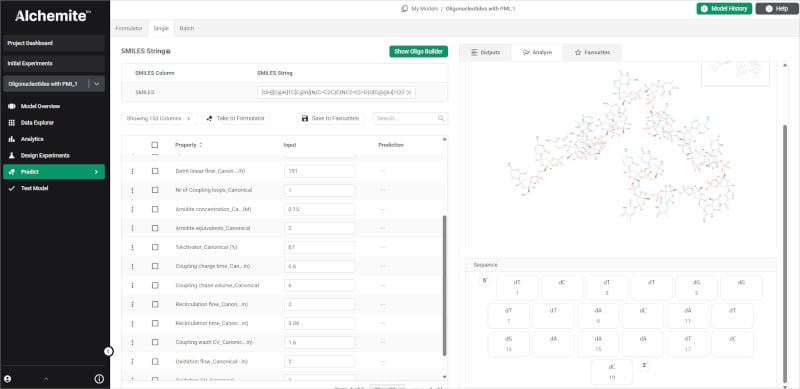
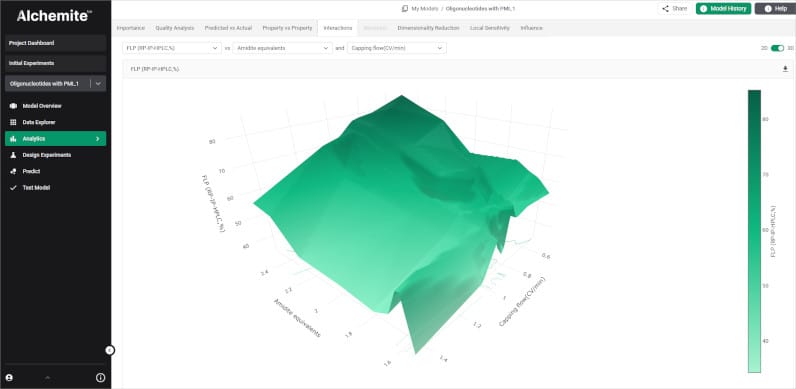
Project results have been impressive, with validation work reducing the experimental burden required to hit defined purity/yield targets, shifting from dozens of experiments to single iteration optimization in many cases. In some examples, critical impurities reduced from around 10% to below 2% and there were crude purity gains of 7-12%. The tool is now available as the Alchemite™ for Oligonucleotide Manufacturing solution and it was demonstrated at a recent Intellegens webinar. If oligonucleotides is your field, you can download a technical paper providing more details of the solution and the project that validated it.
This remains a tricky area, but machine learning is making life for oligos researchers a little easier and bringing the associated dream of new therapies a little closer.